服务简介
Service Introduction表观遗传学是研究基因的核苷酸序列不发生改变的情况下,基因表达可遗传变化的一门遗传学分支学科。DNA甲基化(DNA methylation)、组蛋白修饰(histone modification)、基因组印记(genomic imprinting)、RNA编辑(RNA editing)、基因沉默、核仁显性、休眠转座子激活和性别相关性基因剂量补偿效应等都是典型的表观遗传现象。
表观遗传调控研究技术分为转录前调控研究技术和转录后调控研究技术。转录前调控研究技术主要包括:MSP、BSP、ChIP、EMSA、DNA Pull down、酵母单杂交(Y1H)、Hi-C;转录后调控研究技术主要包括RIP、RNA Pull down 、双萤光素酶报告系统等。
金开瑞致力于ChIP、EMSA、DNA/RNA pull down、RIP、双萤光素酶系统、MSP(甲基化特异性PCR法)、BSP(亚硫酸氢钠处理后测序法)等实验技术服务。除了提供优质的技术服务外,我们还可以为您提供表观遗传学整体项目设计,整体项目实施,整体项目跟踪服务,金开瑞多年的实验和项目管理经验,将助力您表观遗传研究。
表观遗传,选择金开瑞,让您无忧。
|
差异表达分析: |
转录前调控: |
在体水平: |
||
 |
 |
 |
||
|
目标分子筛选、鉴定、定位等 |
 |
分子机制研究 |
 |
功能研究 |
 |
 |
 |
||
|
定位分析: |
转录后调控: |
转录前调控研究技术
ChIP 染色体免疫共沉淀(Chromatin Immunoprecipitation,ChIP)是基于体内分析发展起来的方法,也称结合位点分析法,在过去十年已经成为表观遗传信息研究的主要方法。这项技术帮助研究者判断在细胞核中基因组的某一特定位置会出现何种组蛋白修饰。ChIP 不仅可以检测体内反式因子与DNA 的动态作用,还可以用来研究组蛋白的各种共价修饰与基因表达的关系。近年来,这种技术得到不断的发展和完善。采用结合微阵列技术在染色体基因表达调控区域检查染色体活性,是深入分析癌症、心血管疾病以及中央神经系统紊乱等疾病的主要通路的一种非常有效的工具。
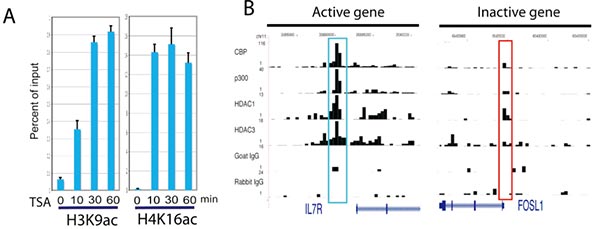
A. Rapid increase of histone acetylation in silent genes caused by HDAC inhibitor treatment.ChIP assays were performed using H3K9ac and H4K16ac antibodies with chromatin from cells treated in the absence or presence of HDAC inhibitors. The ChIP DNA was analyzed using qPCR.
B. The ChIP-Seq signals for the DIPA gene (active) and FOSL1 gene (silent) were displayed.The signals highlighted in blue are not statistically significant as determined using peak-finding programs.
(Wang Z, Zang C, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes [J]. Cell. 2009 Sep 4;138(5):1019-31.)
凝胶迁移或电泳迁移率检测(Electrophoretic Mobility Shift Assay,EMSA)是一种检测蛋白质和DNA 序列相互结合的技术,可用于定性和定量分析。根据实验设计特异性和非特异性探针,当核酸探针与样本蛋白混合孵育时,样本中可以与核酸探针结合的蛋白质与探针形成蛋白-探针复合物。这种复合物由于分子量大,在进行聚丙烯酰胺凝胶电泳时迁移较慢,而没有结合蛋白的探针则较快。孵育的样本在进行聚丙烯酰胺凝胶电泳并转膜后,蛋白-探针复合物会在膜靠前的位置形成一条带,说明有蛋白与目标探针发生互作。目前已用于研究RNA 结合蛋白和特定的RNA 序列的相互作用,是转录因子研究的经典方法。
金开瑞提供EMSA 检测技术服务,帮助您检测DNA 结合蛋白、RNA 结合蛋白、特定的蛋白质,并可进行未知蛋白的鉴定。
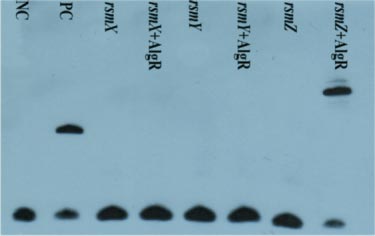
The EMSA results of binding of AlgR to the rsmX/Y/Z promoter sequence.
(Li M, Yan J, Yan Y. The Pseudomonas transcriptional regulator AlgR controls LipA expression via the noncoding RNA RsmZ in Pseudomonas protegens Pf-5 [J]. Biochem Biophys Res Commun. 2017 May 20;487(1):173-180.)
甲基化特异性是指用亚硫酸氢钠处理基因组DNA,未甲基化的胞嘧啶变成尿嘧啶,而甲基化的胞嘧啶不变,然后用3对特异性的引物对所测基因的同一核苷酸序列进行扩增。扩增产物用DNA 琼脂糖凝胶电泳,凝胶扫描观察分析结果。
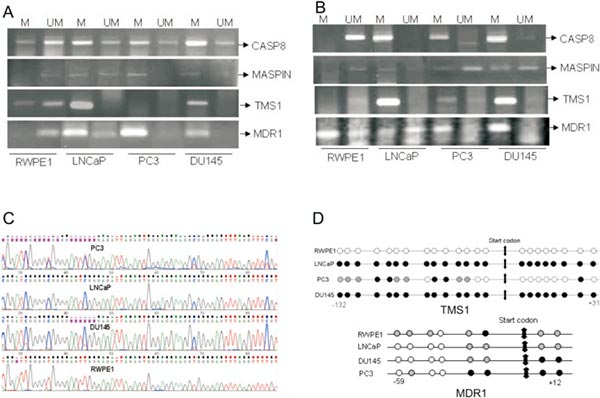
A. Methylation specific PCR of CASP8, MASPIN, TMS1 and MDR1 in prostate cancer cells. DNA from prostate cancer cells were Bisulfite modified using EZ methylation kit (Zymo Research) and then PCR done using methylation specific primers and unmethylation specific primers. Because in the cells sometimes methylated and unmethylated both copies are present, so depending upon the number copies they get amplified.
B. Methylation specific PCR of CASP8, MASPIN, TMS1 and MDR1 in 5-aza-dC (5µM/48hrs) treated prostate cancer cells.
C. Bisulfite sequencing chromatogram of TMS1 gene showing methylation in LNCaP, DU145 and PC3, partial methylation in PC3 and Unmethylation in RWPE1.
D. Diagrammatic representation of methylation pattern of TMS1 and MDR1 gene at different promoter positions in RWPE1, LNCaP, DU145 and PC3. Dark circles (●) represents methylated sites, shadowed circles (An external file that holds a picture, illustration, etc.Object name is nihms159165ig1.jpg) represent partially methylated sites and blank circles (○) represents unmethylated sites in TMS1 and MDR1 promoter region.
(Mishra DK, Chen Z, et al. Global methylation pattern of genes in androgen-sensitive and androgen-independent prostate cancer cells [J]. Mol Cancer Ther. 2010 Jan;9(1):33-45.)
亚硫酸氢盐处理后测序法:这种方法一度被认为是DNA 甲基化分析的金标准。它的过程如下:经过亚硫酸氢盐处理后,用PCR 扩增目的片段,并对PCR 产物进行测序,将序列与未经处理的序列进行比较,判断CpG 位点是否发生甲基化。这种方法可靠,且精确度高,能明确目的片段中每一个CpG 位点的甲基化状态。
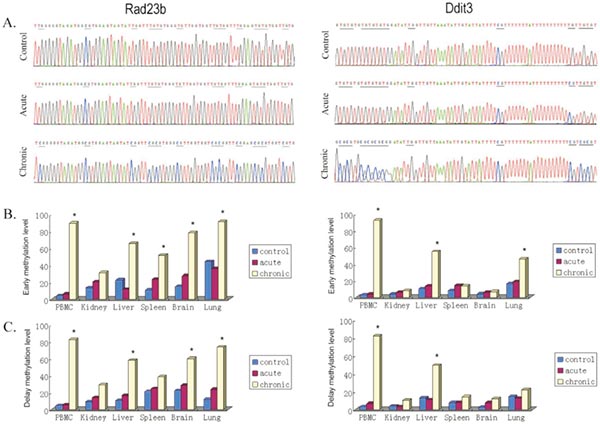
Rad23b and Ddit3 methylation determined by BSP. Bisulfite treated DNA was amplified by PCR with BSP primers. PCR products were cloned into the pUC57 vector, and five clones selected and sequenced from each sample. Methylation level was defined as the ratio of methylated CpG sites in all clones.
A. Typical sequencing results;
B. early effects, tissues were collected 2 h postirradiation;
C. delay effects, 1 month postirradiation. *P,0.05 versus control.
(Wang J, Zhang Y, et al. Genome-wide screen of DNA methylation changes induced by low dose X-ray radiation in mice [J]. PLoS One. 2014 Mar 10;9(3):e90804.)
DNA pull down是用于分析蛋白质与DNA互作的一种分析技术。一般来说,该实验首先需要针对待研究目的基因的调控区域设计并制备特异性探针(以脱硫生物素标记为主流)。同时,制备细胞核提取物;将探针和核提取物共同孵育,DNA 结合蛋白就会和靶向序列特异性结合。然后,通过亲和素磁珠纯化蛋白质DNA 复合物。最后针对获得的蛋白,使用WB 验证或者质谱方式鉴定蛋白质类型。
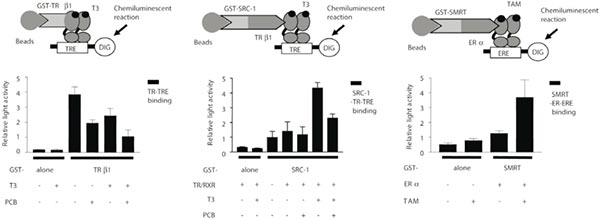
Examples of the liquid chemiluminescent DNA pull-down assay. (A–C) Light emission was measured from the liquid chemiluminescent DNA pull-down assay to indicate binding. The histograms indicate relative light emission. Data are shown as means ± x001D. (A) Effect of polychlorinated biphenyl (PCB) on thyroid hormone receptor β1 (TRβ1)-thyroid hormone response element (TRE) binding. Glutathione S-transferase (GST)-TRβ1 was incubated with digoxigenin (DIG)-labeled TRE with or without T3 and/or PCB. (B) Effect of PCB on steroid receptor coactivator-1 (SRC-1)-TRβ1-TRE binding. GST-SRC-1 was incubated with in vitro-translated TRβ1 and retinoid X receptor (RXR), then SRC-1-TRβ1/RXR complex was incubated with DIG-labeled TRE with or without T3 and/or PCB for 1 h at room temperature. (C) Effect of tamoxifen on silent mediator for retinoid and thyroid hormone (SMRT)-estrogen receptor α(ERα )-estrogen response element (ERE) binding. GST-SMRT was incubated with ERα, then SMRT-ERα complex was incubated with DIG-labeled ERE with or without 17β-estradiol (E2) and/or tamoxifen (TAM). After detection reaction, light emission was measured to indicate binding as described above. Data are shown as means ±SEM.
(Iwasaki T, Miyazaki W, et al. Liquid chemiluminescent DNA pull-down assay to measure nuclear receptor-DNA binding in solution [J]. Biotechniques. 2008 Oct;45(4):445-8.)
将提取的样品RNA 逆转录成单链cDNA,经长距离聚合酶链反应扩增得到双链cDNA,再经CHROMA SPIN TE-400 柱子纯化后与酵母表达载体pGADT7-Rec 线性质粒共转至Y1HGold [pBait-AbAi] 酵母感受态,经SD/-Leu/AbA 缺陷培养基筛选出与诱饵序列相互作用的猎物蛋白。
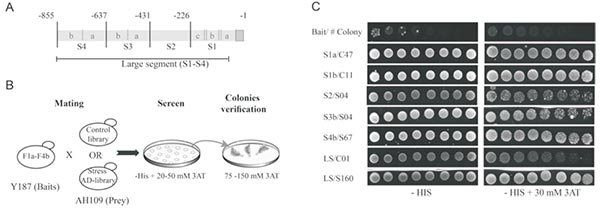
Description of the Y1H screens approach conducted with Zat12 promoter segments as baits.
(Ben Daniel BH, Cattan E, et al. Identification of novel transcriptional regulators of Zat12 using comprehensive yeast one-hybrid screens [J]. Physiol Plant. 2016 Aug;157(4):422-41.)
Hi-C (High-through chromosome conformation capture) 是以整个细胞核为研究对象,利用高通量测序技术,结合生物信息分析方法,研究全基因组范围内整个染色质DNA 在空间位置上的关系,获得高分辨率的染色质调控元件相互作用图谱。Hi-C 可以与RNA-Seq、ChIP-Seq 等数据进行联合分析,从基因调控网络和表观遗传网络来阐述生物体性状形成的相关机制。
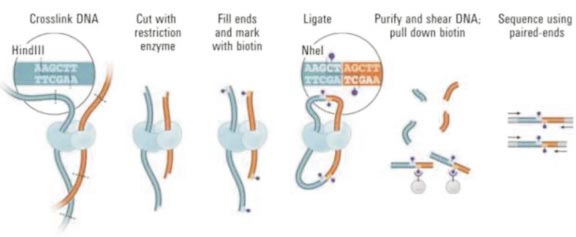
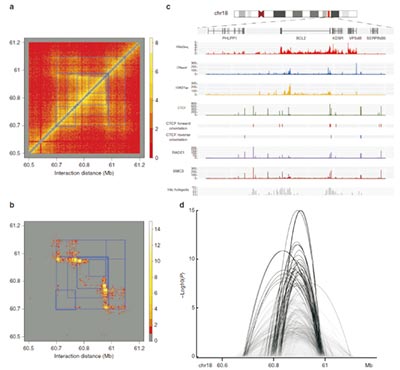
Figure. HiC-DC identifies identifies interactions associated with regulatory and structural elements at the sub-TAD level.
a. Raw Hi-C count matrix from the Rao et al.11 GM12878 data set for aB700 kb region including the BLC2 locus. Sub-TAD regions as called by Rao et al.11 are shown as blue squares.
b. Significant interactions (–log10 P values) for the same region as estimated by HiC-DC.
c. Epigenomic tracks for GM12878 for the same region, showing DNase I hypersensitive sites, H3K27ac, ChIP-seq for components of the cohesin complex, and Hi-C hotspots as estimated by HiC-DC.
d. Sashimi plot depiction of significant interactions called by HiC-DC, showing chromatin looping between hotspots associated with regulatory and structural elements.
(Carty M, Zamparo L, et al. An integrated model for detecting significant chromatin interactions from high-resolution Hi-C data [J]. Nat Commun. 2017 May 17;8:15454.)
转录后调控研究技术
RIP 技术(RNA Binding Protein Immunoprecipitation Assay,RNA 结合蛋白免疫沉淀)主要是运用针对目标蛋白的抗体把相应的RNA-蛋白复合物沉淀下来,然后经过分离纯化就可以对结合在复合物上的RNA 进行q-PCR 验证或者测序分析。RIP 是研究细胞内RNA 与蛋白结合情况的技术,是了解转录后调控网络动态过程的有力工具,可以帮助我们发现miRNA 的调节靶点。
根据客户需求,金开瑞可以提供RNA/蛋白、RNA/RNA 互作验证RIP 技术服务。
● 使用目的蛋白寻找验证RNA(蛋白/RNA互作):
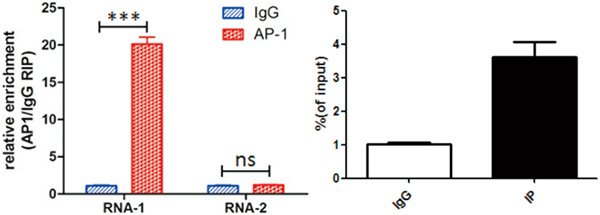
图. AP-1蛋白与RNA的相互作用RIP
● 使用目的lncRNA寻找验证miRNA(RNA/RNA互作):
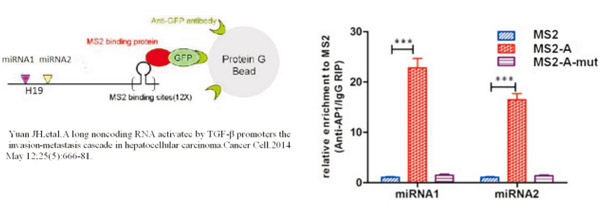
图. MS2- RIP实验原理图(表达Lnc-A的质粒富集miRNA1miRNA2的效率比不表达Lnc-A的MS2组明显提高。通过对照,说明MS2-A可以与miRNA1和miRNA2相互结合)
使用体外转录法标记生物素RNA 探针,然后与胞浆蛋白提取液孵育,形成RNA-蛋白质复合物。该复合物可与链霉亲和素标记的磁珠结合,从而与孵育液中的其他成分分离。复合物洗脱后,通过WB 实验检测特定的RNA 结合蛋白是否与RNA 相互作用。蛋白质与RNA 的相互作用是许多细胞功能的核心,如蛋白质合成、mRNA 组装、病毒复制、细胞发育调控等。若待检测目的蛋白明确,选择WB 鉴定;若不明确,则可选择质谱鉴定。
金开瑞现提供RNA pull down 检测技术服务,使用特异性二抗进行检测,能有效避免抗体重链对结果的信号干扰,配套先进的质谱仪,能显著提升检测效果。
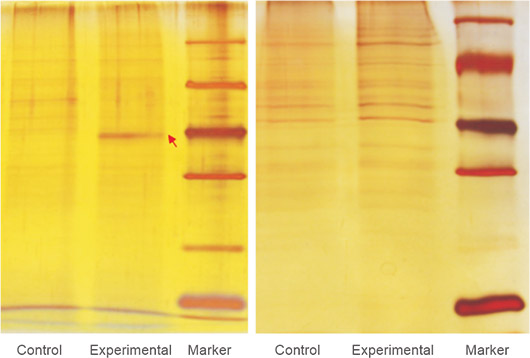
图. pull down下来的蛋白银染图。根据胶上条带的差异情况和实验目的,选择质谱或者WB鉴定pull down下来的蛋白

图. SDS-PAGE电泳后质谱鉴定
双萤光素酶报告基因用于实验系统中作相关的或成比例的检测,通常一个报告基因作为内对照,使另一个报告基因的检测均一化。检测基因表达时双报告基因通常用来瞬时转染培养细胞,带有实验报告基因的载体共转染带有不同的报告基因作为对照的第二个载体。通常实验报告基因偶联到调控的启动子,研究调控基因的结构和生理基础。报告基因表达活力的相对改变与偶联调控启动子转录活力的改变相关,偶联到组成型启动子的第二个报告基因,提供转录活力的内对照,使测试不被实验条件变化所干扰。通过这种方法, 可减少内在的变化因素所削弱的实验准确性, 如培养细胞的数目和活力的差别, 细胞转染和裂解的效率。理想的双报告基因方法应该使用户能够以萤火虫萤光素酶所具有的速度,灵敏和线性范围对同一样品中的两个报告基因同时测定。
金开瑞应用Promega 公司的双萤光素酶报告基因检测(DLR)系统,提供DLR 检测服务。
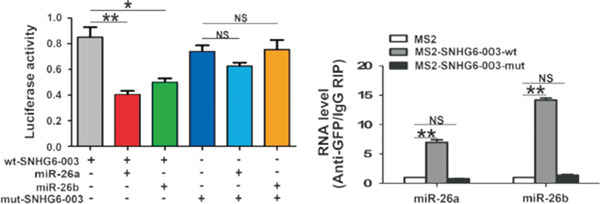
图. 双萤光素酶和RIP验证lncRNA可以与miR-26a结合
(C Cao, Zhang T, Zhang D, et al.The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma [J]. Oncogene. 2017 Feb 23;36(8):1112-1122. )











